7.3. Adding a CSV file format to spectrum read/write
In this section we're going to walk through the problem of writing a new formatter and making it known to the SpecTcl sread and swrite commands.
The format will be a simple CSV file format. For 1-d spectra, a spectrum contains a single line of comma separated channel values. For 2-d spectra, each line is a scan line of the spectrum. For simplicity(?) no metadata will be stored or restored. Read will only create 1-d and 2-d snapshot spectra.
Here's the header to our CSVSpectrumFormatter
class:
Example 7-1. CSVSpectrumFormatter header
#ifndef CSVSPECTRUMFORMATTER_H
#define CSVSPECTRUMFORMATTER_H
#include <SpectrumFormatter.h>
class CSVSpectrumFormatter : public CSpectrumFormatter
{
public:
virtual CSpectrum* Read (STD(istream)& rStream,
ParameterDictionary& rDict) ;
virtual void Write (STD(ostream)& rStream, CSpectrum& rSpectrum,
ParameterDictionary& rDict);
};
#endif
Let's get our feet wet with the Write operation.
Here's how we're going to do that:
Get the dimensionality of the spectrum (1 or two)
From the dimensionality of the spectrum construct the number of scan lines to output, note that for 1-d spectra this is one.
Iterate over each scanline, writing comma separated values terminating the scanline with a \n.
Let's look at the the implementation. First, we introduce the
simple utility method writeScanline:
The parameters are pretty clear, y
is the y coordinate of the scanline and n
the number of channels on each scanline. Here is the implementation
of this helper:
Example 7-2.
Implementation of writeScanline method.
#include "CSVSpectrumFormatter.h"
#include <Spectrum.h>
void
CSVSpectrumFormatter::writeScanline(std::ostream& rstream, CSpectrum& spec, unsigned y, unsigned n)
{
UInt_t indices[] = {0, y};  for (int i = 0; i < n; i++) {
ULong_t ch = spec[indices];
for (int i = 0; i < n; i++) {
ULong_t ch = spec[indices];  char delim = ',';
if (indices[0] == (n-1)) {
char delim = ',';
if (indices[0] == (n-1)) {  delim = '\n';
}
rstream << ch << delim;
indices[0]++;
}
}
delim = '\n';
}
rstream << ch << delim;
indices[0]++;
}
}
-
This array will be an array of indices into the spectrum.
CSpectrumimplements anoperator[]but, due to the need to handle a variable number of indices, it takes an array of indices rather than a single index. 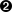
-
The indexing operator of
CSpectrumis supposed to return the value of the channel at the coordinates provided by the index array. Note that if this is a 1-d spectrum the second index is ignored. 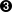
- Each channel output wil be followed by a delimeter. For all but the last item on the scanline, the delimeter is , for the last item, the delimeter is a newline character.
Next we can write the Write method in terms
of this utility:
Example 7-3. CSVSpectrumFormatter::Write implementation
void
CSVSpectrumFormatter::Write (STD(ostream)& rStream, CSpectrum& rSpectrum,
ParameterDictionary& rDict)
{
UInt_t nDims = rSpectrum.Dimensionality(); 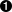 UInt_t xDim = rSpectrum.Dimension(0);
UInt_t xDim = rSpectrum.Dimension(0); 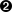 UInt_t yDim;
if (nDims == 1) {
yDim = 1;
UInt_t yDim;
if (nDims == 1) {
yDim = 1;  } else {
yDim = rSpectrum.Dimension(1);
} else {
yDim = rSpectrum.Dimension(1);  }
for (int i = 0; i < yDim; i++) {
writeScanline(rStream, rSpectrum, i, xDim);
}
for (int i = 0; i < yDim; i++) {
writeScanline(rStream, rSpectrum, i, xDim);  }
}
}
}
- Fetches the number of dimensions the spectrum has. This is one or two. Note we must use this rather than getting the parameter count or Axis count as these can be misleading for some spectrum types.
- Retrieves the number of channels on the X axis. All spectra have at least an X axis.
-
yDimis the number of scanlines we'll need to write. If the spectrum has only one dimension, we're only writing a single scanline. 
- On the other hand, if the spectrum has two dimensions, the number of channels in Y determines the number of scan lines to write.
- This loop writes all the scanlines we need to write.
Reading in a spectrum in CSV form requires that we have some way to parse a CSV file. We are going to follow the time honored practice of using code someone already wrote to do this. I am a firm believer that good programmers are lazy thieves.
The code we're going to use to parse the CSV files is sample code at: http://www.zedwood.com/article/cpp-csv-parser. We only need the last function on that page. We'll incorporate it as a utility method in our spectrum formatter;
You can look at the citation above to see the actual code for this method. For the sake of brevity we're going to treat it as a black box and not show the code here.
What we need to do to read back spectra is to:
Read a scanline as CSV converting the strings to numbers. It's an error if all strings in the CSV decode don't parse to integers.
The X dimension of the spectrum is determined by the size of this current scanline.
If there are no more scanlines, this is a 1-d spectrum create it as a snapshot and store the scanline in the spectrum.
If there are additional scanlines, build up the 2-d array scanline by scanline. The number of scanlines determines the Y dimension of the spectrum.
Create the 2-d snapshot spectrum and save the scanlines in it.
We're going to accumulate the data in a std::vector< std::vector < unsigned > >. Each element of the outer vector is a scan line.
There are a few things for which it's worth providing some utility methods:
CSpectrum* create1DSpectrum(int nx);
CSpectrum* create2DSpectrum(int nx, int ny);
CParameter* dummyParameter(SpecTcl& api);
std::string uniqueName(const char* basename, SpecTcl& api);
create1DSpectrumCreates and returns a pointer to a uniquely named 1-d spectrum using a dummy parameter.
create2DSpectrumCreates and returns a pointer to a uniquely named 2-d spectrum using a dummy parameter on both axes.
dummyParameterIf a dummy parameter named _csv_dummy_param exists it is returned, otherwise one is created and that is returned. This is necessary because spectrum objects in SpecTcl must have parameters. We'll use this one to make it clear to people listing spectra that the parameter is meaningless from the point of view of other analysis,.
uniqueNameFinds a spectrum name not yet in use for names like basename (first one tried) and basename_integer.
These methods are relatively simple:
CSpectrum*
CSVSpectrumFormatter::create1DSpectrum(int nx)
{
SpecTcl& api(*(SpecTcl::getInstance()));
CParameter* pDummyParam = dummyParameter(api);
std::string spectrumName = uniqueName("csvspectrum", api);
return api.Create1D(spectrumName, keLong, *pDummyParam, nx);
}
CSpectrum*
CSVSpectrumFormatter::create2DSpectrum(int nx, int ny)
{
SpecTcl& api(*(SpecTcl::getInstance()));
CParameter* pDummyParam = dummyParameter(api);
std::string spectrumName = uniqueName("csvspectrum", api);
return api.Create2D(spectrumName, keLong, *pDummyParam, *pDummyParam, nx, ny);
}
CParameter*
CSVSpectrumFormatter::dummyParameter(SpecTcl& api)
{
CParameter* result = api.FindParameter("_csv_dummy_param");
if (!result) {
result = api.AddParameter("_csv_dummy_param", api.AssignParameterId(), "");
}
return result;
}
std::string
CSVSpectrumFormatter::uniqueName(const char* baseName, SpecTcl& api)
{
std::string result = baseName;
int index = 0;
while(1) {
if(!api.FindSpectrum(result)) return result;
std::stringstream s;
s << baseName << "_" << index;
result = s.str();
index++;
}
return result;
}
The only tricky thing is how unique name
loops trying to find spectra that match candidaten ames.
If there is no match, a unique name has been found and is returned.
Adjusting the name and index at the bottom of the while loop allows
for the baseName to be tried without any
adornments.
Armed with these utilities, let's write the Read
method:
Example 7-4. CSVSpectrumFormatter::Read implementation
CSpectrum*
CSVSpectrumFormatter::Read (STD(istream)& rStream,
ParameterDictionary& rDict)
{
std::vector<std::vector<unsigned> > scanlines;  std::vector<std::string> csvline;
std::vector<std::string> csvline;  while(!rStream.eof()) {
csvline.clear();
csvline = csv_read_row(rStream);
while(!rStream.eof()) {
csvline.clear();
csvline = csv_read_row(rStream);  if (csvline.size()) {
if (csvline.size()) {  std::vector<unsigned> line;
for (int i = 0; i < csvline.size(); i++) {
char* endptr;
unsigned v = strtoul(csvline[i].c_str(), &endptr, 0);
std::vector<unsigned> line;
for (int i = 0; i < csvline.size(); i++) {
char* endptr;
unsigned v = strtoul(csvline[i].c_str(), &endptr, 0);  if (endptr == csvline[i].c_str()) {
throw std::string("Failed conversion to integer in CSVSpectrumFormatter::Read");
}
line.push_back(v);
if (endptr == csvline[i].c_str()) {
throw std::string("Failed conversion to integer in CSVSpectrumFormatter::Read");
}
line.push_back(v);  }
scanlines.push_back(line);
}
scanlines.push_back(line);  }
}
CSpectrum* pSpectrum(0);
}
}
CSpectrum* pSpectrum(0);
 if (scanlines.size() == 1) {
pSpectrum = create1DSpectrum(scanlines[0].size());
} else {
pSpectrum = create2DSpectrum(scanlines[0].size(), scanlines.size());
}
UInt_t indices[2];
for (int y = 0; y < scanlines.size(); y++) {
if (scanlines.size() == 1) {
pSpectrum = create1DSpectrum(scanlines[0].size());
} else {
pSpectrum = create2DSpectrum(scanlines[0].size(), scanlines.size());
}
UInt_t indices[2];
for (int y = 0; y < scanlines.size(); y++) {  for (int x = 0; x < scanlines[y].size(); x++) {
UInt_t indices[] = {x, y};
pSpectrum->set(indices, scanlines[y][x]);
}
}
return new CSnapshotSpectrum(*pSpectrum);
for (int x = 0; x < scanlines[y].size(); x++) {
UInt_t indices[] = {x, y};
pSpectrum->set(indices, scanlines[y][x]);
}
}
return new CSnapshotSpectrum(*pSpectrum);  }
}
-
scanlineswill hold all of the spectrum channels read fromt he file. We can't just build the spectrum into aCSpectrumobject because we don't know how to declare that object until we have read in all the scanlines. This variable consists of a vector whose elements are the values of channels in one scanline. A scanline is just the channels in a spectrum with fixed y coordinate.
-
csvlinewill hold the value of one scanline read by the CSV decoding method. Note that scanline is a vector of strings which must then be converted into a vector of unsigned values. 
- This line decodes a line fromt he CSV file. This will be done repeatedly until we have an endfile condition on the input file. This format, does not support packing several spectra into a single file.
-
We only have to decode the scanline into integers and
store it in
scanlinesif there are entries decoded from thel line. Two reasons thecsvlinemight be empty are blank lines embedded in the file (by a creator other than us) or blank lines at the end of the file prior to the EOF condition. 
-
This call to
strtoulattempts to decode a string in a cell from the line into an unsigned value.endptr, on success, points after the decoded string. On failure,endptrwill point to the beginning of the string. Any failure indicates this is not a valid spectrum file. We flag this by throwing an exception. 
-
If the cell was converted successfully, the unsigned value
is pushed back into the
linevector in which the integer values of the scanline are being accumulated. 
-
Once a scaneline has been decoded it is pushed back into the
scanlinesvector. 
-
This section of code creates the spectrum into which the
data in
scanlineswill be stored. If the file only had a single scanline, the data are for a 1-d spectrum. Otherwise the data are for a 2-d spectrum. With this simple file format we can't distinguish between anything other than 2-d and 1-d spectra. A summary spectrum, for example, looks like a 2-d spectrum. This doesn't matter since we're not going to hook the spectrum up to be incremented.
-
Fills the channels of the spectrrum with the data in
scanlines. We nkow the data will fit because we used the dimensionality ofscanlinesas the spectrum dimensions with creating it. 
- The final spectrum is wrapped by a snapshot spectrum. While we used a nonesnsical parameter to construct the spectrum, this makes doubly sure SpecTcl won't try to connect it to the histogrammer.
Note that the sread command may wrap this in a snapshot spectrum but this wrapping ensures that even if
-nosnapshotis used, the spectrum will be a snapshot.
Having written this extension, the only thing left to do is
to make SpecTcl aware of this. The API method
AddSpectrumFormatter can do this.
Probably the best place to do this is in
MySpecTclApp::CreateHistogrammer.